In the previous post we briefly introduced the Avida software platform for the study of evolution. Here, we will learn to work with parasites within this computational tool. Parasitic digital organisms are almost identical to the hosts, and as such they self-replicate by copying their genome instruction by instruction into a new memory space. But they operate inside hosts, stealing CPU cycles from them to execute their own genome’s instructions and, hence, reduce their host fitness.
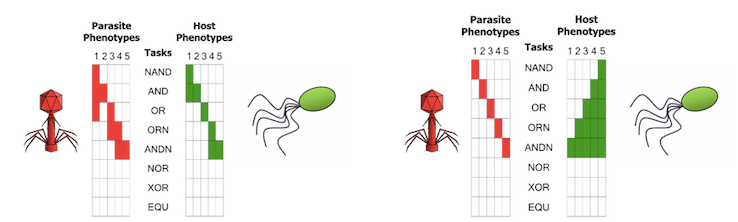
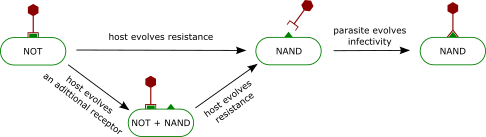
Digital coevolution between hosts and parasites resembles, at some level, the coevolutionary dynamics among bacteria and phages (ie.,. viruses that infect bacteria). On the one hand, bacteria must have receptors on their surface in order to import resources from their environment. On the other hand, phages must attach to those receptors in order to infect bacteria. Therefore, a trade-off exists between having receptorsf or obtaining nutrients and being susceptible to phages.
Coevolutionary dynamics results from bacteria evolving phage resistance by changing their surface receptors, and from phages countering bacteria resistance by altering their tail fibres to attach to the novel receptor.
Analogously, digital hosts must compute logic operations to consume resources and thus replicate, but those traits leave them susceptible to infection by digital parasites.
Installation.
We need first to install git (see how to do it from JuptyterLab).
Then, from the devosoft/avida development account on github
we get the repository after setting-up git from the command line:
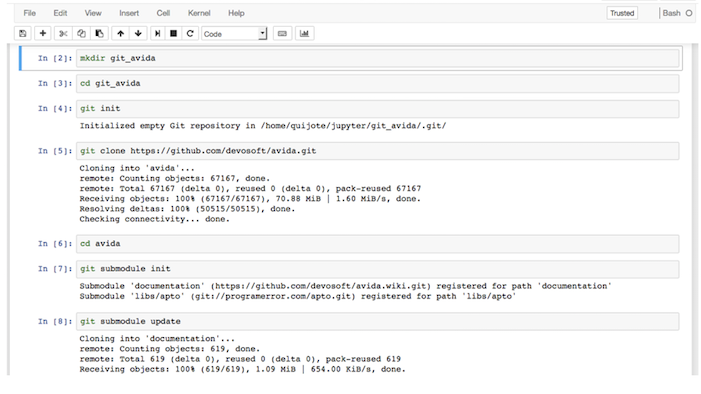
mkdir git_avida # create folder
cd git_avida # go to folder
git init # initialize git repository (it creates a subfolder named .git)
git clone https://github.com/devosoft/avida # get repository
and, finally, we follow the instructions:
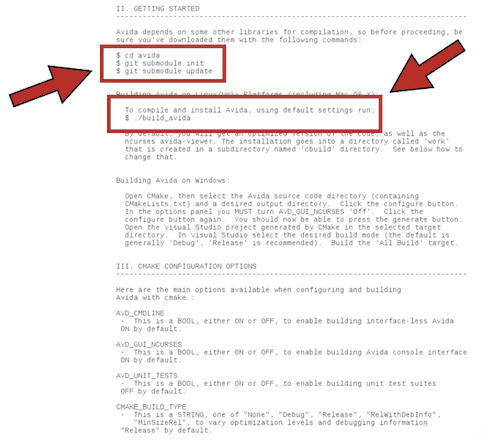
cd avida
git submodule init
git submodule update
We then compile and install Avida:
./build_avida
Of course, we can do it from JuptyterLab as well:
Configuration.
After a successful installation, we will have the following directory structure:
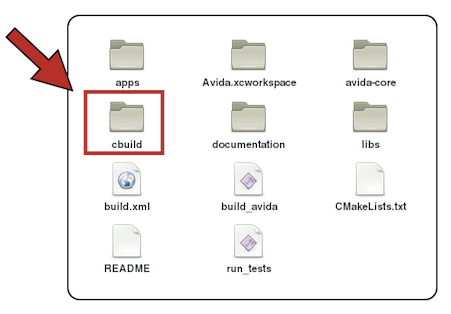
We go to cbuild and then to work
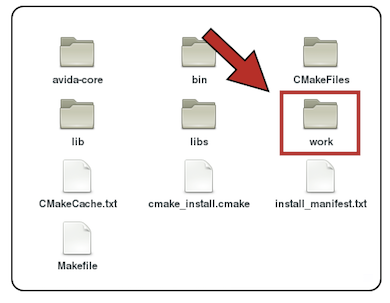
The work folder contains the configuration files *.cfg and the executable file avida.
The avida.cfg file is the main configuration file for designing an experiment.
It's a text file containing many options along with a description of what each one does and what its parameters are.
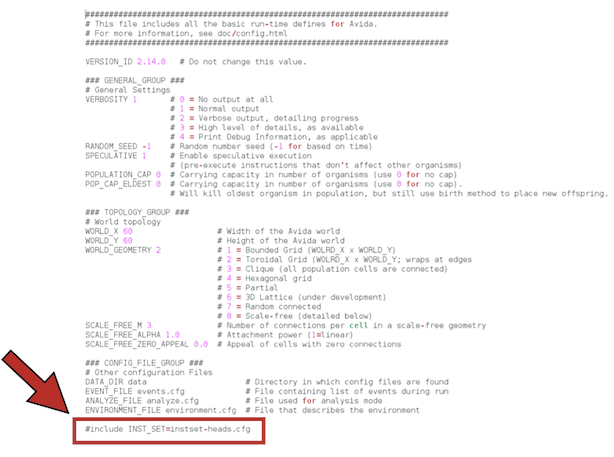
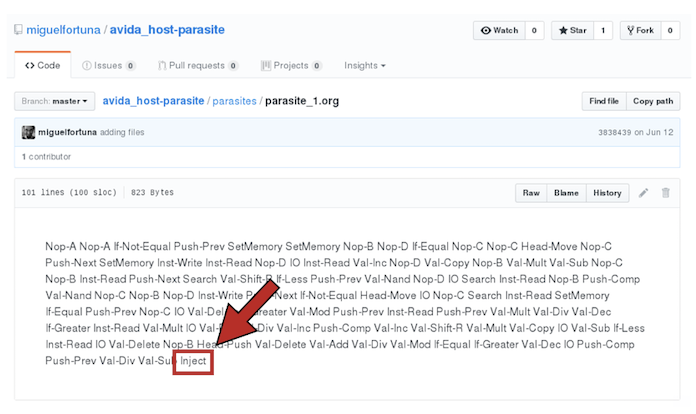
In order for Avida to work with parasites, we need to change the instruction set that is going to be used in the experiment. That is, the genetic language for digital organisms (analogous to the four nucleotides for biological organisms). By default, the genome of a digital organism is made of instructions taken from the set of 26 instructions specified in the file instset-heads.cfg (you can also see the instset-heads-sex.cfg file in the folder, which is used in experiments in which recombination between organisms is allowed). Parasites and their hosts work under a different instruction set, instset-transsmt.cfg. This 33-instruction set contains the instruction Inject that, when executed, is used for the parasite to infect its host. Do not forget to change this! It's the only option we're going to modify by editing the avida.cfg file directly. We will set the configuration options when running Avida from the command line.
There are many ways for a digital organism to mutate,

many demographic parameters to be set,

and many options for them to interact with the environment.
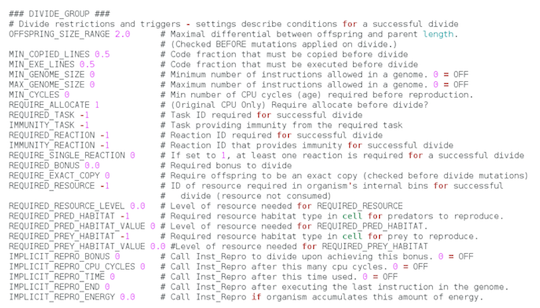
The PARASITE GROUP options are the ones required for setting up parasites.

You can get more information on how parasites work in Avida from his developer, Luis Zaman.
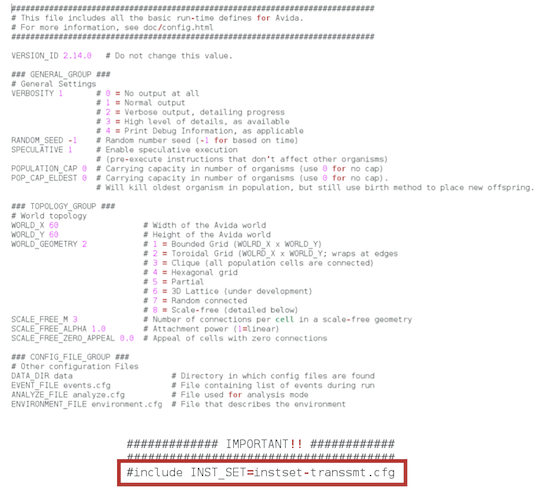
If the instset-transsmt.cfg file provided by default looks like the one below,

please, replace it by this one (the instruction Inject should be there, as well as Divide-Erasethat is used to produce offspring under this instruction set):
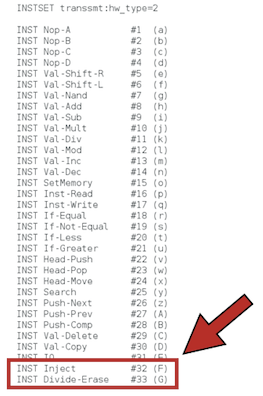
You can see the meaning of the 33 instructions contained in the instset-transsmt.cfg from our repository.
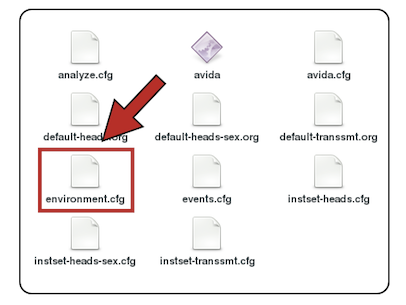
The environment.cfg file contains the options that will allow digital organisms to get extra CPU cycles (and hence self-replicate faster) when computing Boolean functions.
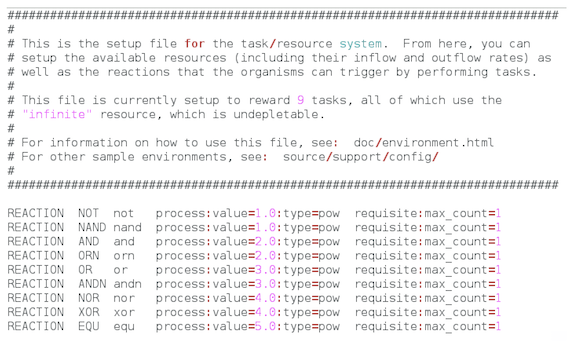
We will replace the content of the default environment.cfg file as follows:
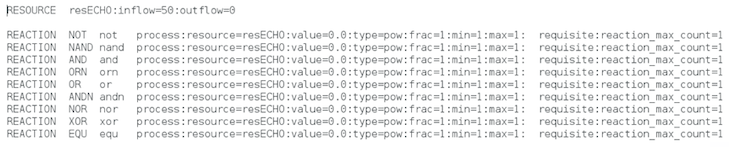
We specify a single resource that will limit population size (first line). Parasites and hosts will take a unit of resource (if available) the first time they compute a Boolean function (any of the 9 functions defined here: NOT, NAND, AND, ORN, OR, ANDN, NOR, XOR, and EQU) by manipulating—with the instructions they may harbor in their genomes—32-binay numbers that the take from the environment. If they do not perform any Boolean function, or if there are not resources available in the environment, the digital organisms will fail when producing offspring.
We do not reward digital organisms with extra CPU cycles for computing Boolean functions. Then, there are no fitness differences between organisms encoding different phenotypes (i.e., computing different Boolean functions): the only selective pressure will come from the antagonistic interactions between hosts and parasites.
The envents.cfg file contains many options to specify the kind of data we want to get from the experiment and when we want them. Below, you can see an example that illustrates many more options that the one we will use here.

The content of our envents.cfg file will be the following:
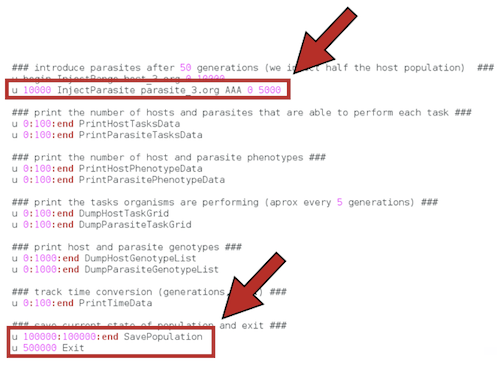
A host population will grow and evolve, from a single ancestor, in the absence of parasites until a single parasite ancestor is introduced at time 5000 (after approximately 100 host generations, for this particular host ancestor). The experiment will last until time 500000 (approximately, 10000 host generations).
You can choose any of these 30 hosts and 15 parasites as ancestors to perform your our coevolutionary experiments:
The genome of one of these hosts looks like (100 instructions):
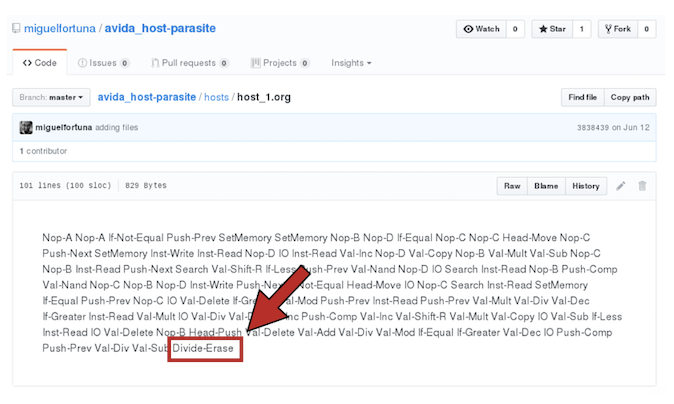
Same for parasites:
Execution.
We run Avida from the command line, or from a bash notebook in JupyterLab, as:
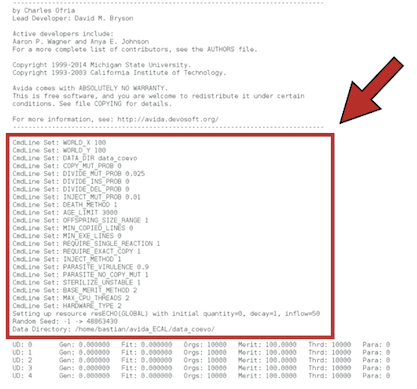
./avida -set DATA_DIR data_coevo -set WORLD_X 100 -set WORLD_Y 100 -set COPY_MUT_PROB 0 -set DIVIDE_INS_PROB 0 -set DIVIDE_DEL_PROB 0 -set OFFSPRING_SIZE_RANGE 1 -set MIN_COPIED_LINES 0 -set MIN_EXE_LINES 0 -set REQUIRE_EXACT_COPY 1 -set STERILIZE_UNSTABLE 1 -set HARDWARE_TYPE 2 -set BASE_MERIT_METHOD 2 -set DEATH_METHOD 1 -set AGE_LIMIT 3000 -set MAX_CPU_THREADS 2 -set PARASITE_VIRULENCE 0.9 -set PARASITE_NO_COPY_MUT 1 -set INJECT_METHOD 1 -set REQUIRE_SINGLE_REACTION 1 -set DIVIDE_MUT_PROB 0.025 -set INJECT_MUT_PROB 0.01
We have modified many options of the configuration file avida.cfg by adding set - option when calling Avida.
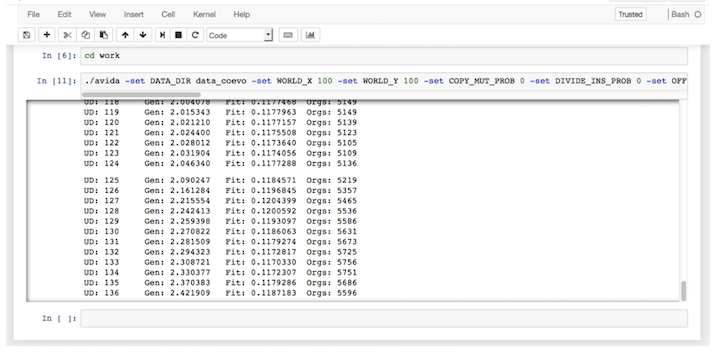
You will see the following output after running the above command in JupyterLab
or in a terminal:
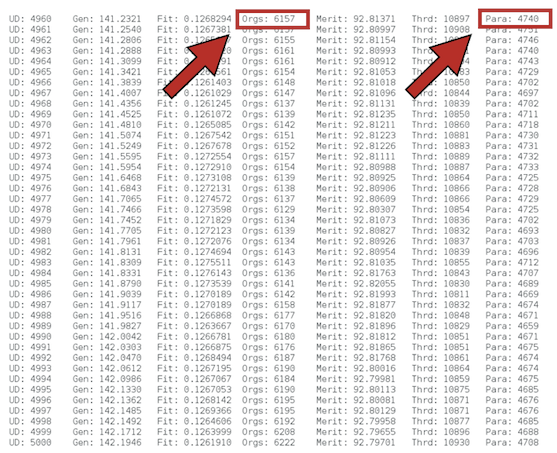
The total number of hosts Orgs: and parasites Para: will change over time as a consequence of the ecological and coevolutionary dyamics.
One the experiment is over, we will ge the following directory structure:
We briefly explain what those files tell us. We start with the file grid_task_hosts.500000.dat

Each number represent the phenotype (i.e., a unique combination of the Boolean functions that a digital organism performed) of the host organism that was living at that time (i.e., 500000, that is, the end of the experiment) in that memory cell within the 100x100 grid (defined by the WORLD_X and WORLD_Y options).
The number -1 indicates that that particular memory cell was empty at that time (i.e., there was no organism living there).
The phenotype of the organism living in a particular memory cell can be obtained by converting the number into the vector of the Boolean functions as (e.g., 4):

Same for the file grid_task_parasite.500000.dat Here, the parasite lives in the same memory cell where its host is living. It cannot be located in an empty memory cell: -1 in the grid_task_hosts.500000.dat

The file host_genome_list.500000.dat contains the genomes of each host organism whose phenotype was described above.

Note that the genomes of all host organisms contain the instruction G (i.e., the one required to produce an offspring Divide-Erase).
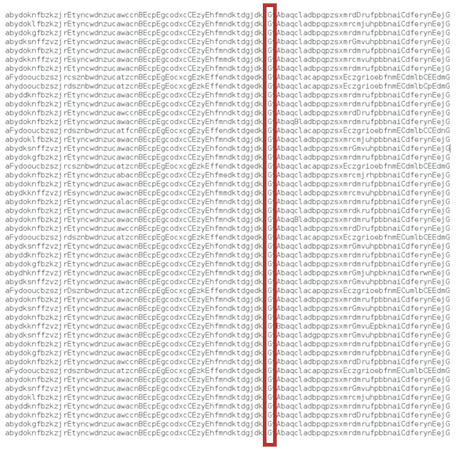
Same for the parasite_genome_list.500000.dat file. Parasite genomes are shorter than host genomes (for this particular experiment).
Here, all parasite's genomes contain the instruction F (the one required to infect a host Inject).

The host_tasks.dat file contains the number of host organisms performing each Boolean function. Note that this is not the number of hosts whose genomes encode a distinct phenotype (i.e., a host organism can perform more than one Boolean function).

Same for the parasite_tasks.dat file. Remember that parasites were introduced at time 5000, that's why we do not see any parasite in the screenshot yet.

Finally the time.dat file contains the mapping between updates (i.e., the unit of time in Avida) and the corresponding number of generations. The time required for an organism to produce an offspring (called in Avida gestation time) depends on the genome, which is continually evolving).

The information provided in those files is enough for generating figures like this one (and movies; see the next post):
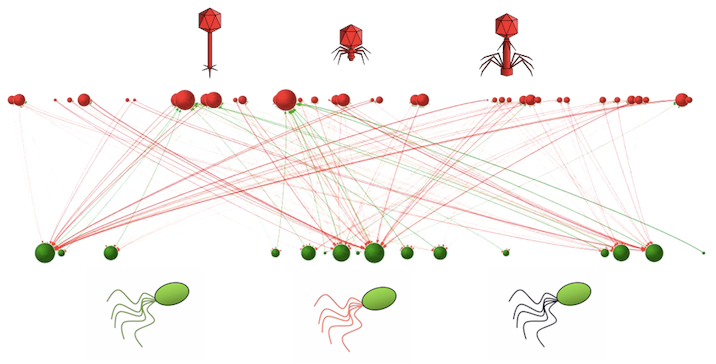
You can take a look at our paper on the role of exaptations in shaping antagonistic host-parasite networks to learn more about setting and running coevolutionary experiments in silico.