The lack of a clearly defined vocabulary makes biologists feel reluctant to embrace the field of digital evolution. The Ontology for Avida (OntoAvida) project is developing an integrated vocabulary for the description of the most widely used computational approach for performing experimental evolution on digital organisms (i.e., self-replicating computer programs that evolve within a user-defined computational environment).
In this post, I show briefly how this project has started.
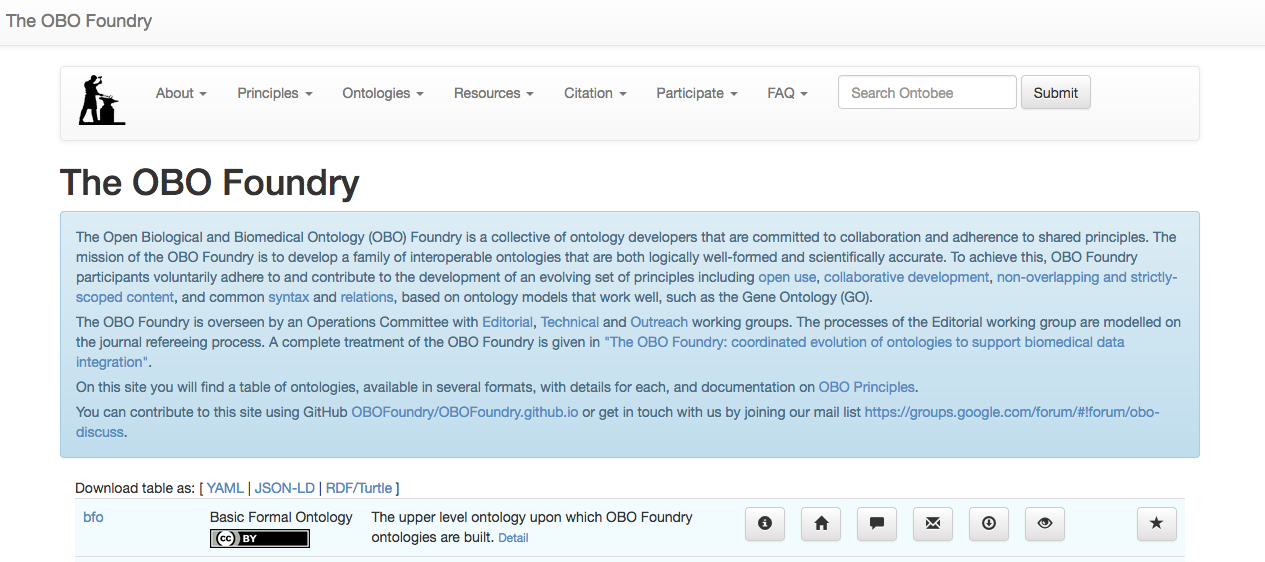
The Ontology Development Kit (ODK), from the OBO Foundry, provides a way of creating an ontology project ready for pushing to GitHub. Just to illustrate the goal of the OBO Foundry, in this site you will find a table of ontologies, available in several formats, with details for each, and documentation on OBO Principles. Please, take a look at some of them and you realize the huge effort of ontology developers to add semantics to the biomedical and biological scientific fields.
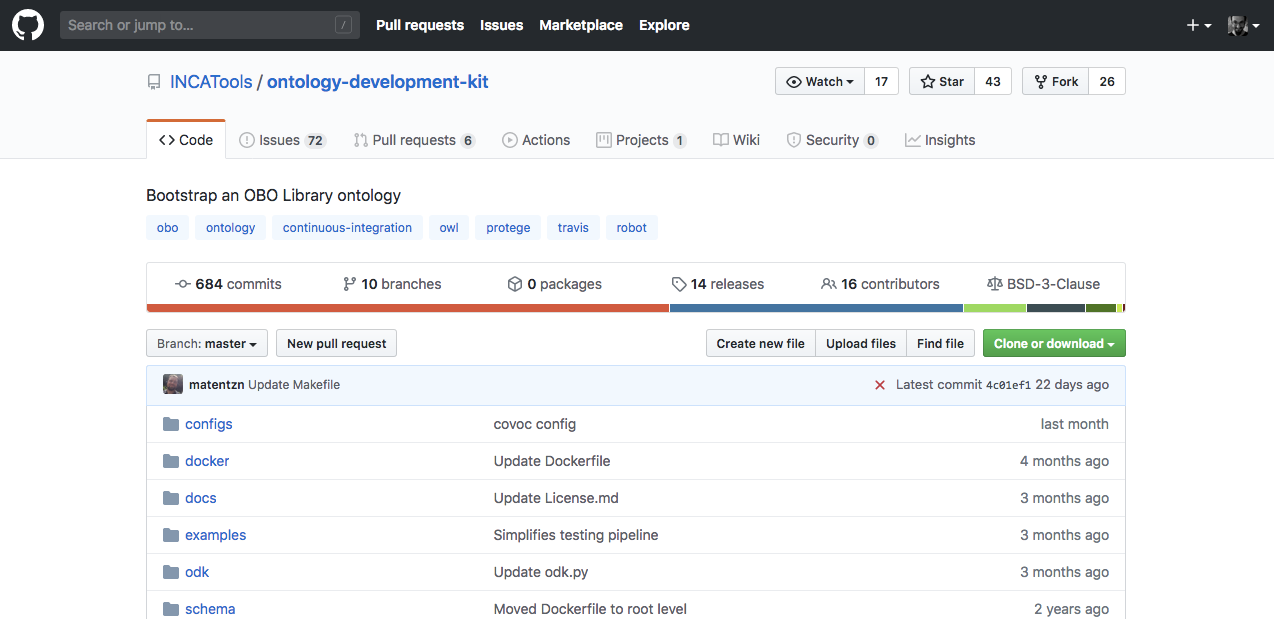
We will use Docker to install the Ontology Development Kit from the GitHub repository.
We first create a new folder for storing our ontology:
mkdir ontology@avida
Then, we pull the software to the newly created folder:
cd ontology@avida
docker pull obolibrary/odkfull
Next, we download the wrapper script that will configure the source tree directory for the ontology:
wget seed-via-docker.sh
We need to make this script executable by changing the permissions and by allowing it to access the Docker sock:
chmod 755 seed-via-docker.sh
chmod 777 /var/run/docker.sock
We customize the project.yaml file that was downloaded along the ODK to the ontology@avida folder (i.e., edit the file using, for example, the nano editor), by adding the following:
id: ontoavida
title: OntoAvida
github_org: fortunalab
repo: ontoavida
Then, we run the wrapper script (make sure you have Docker running):
./seed-via-docker.sh -C project.yaml
After changing the permissions to access the source tree directory:
sudo chown -R yourusername:yourusername target
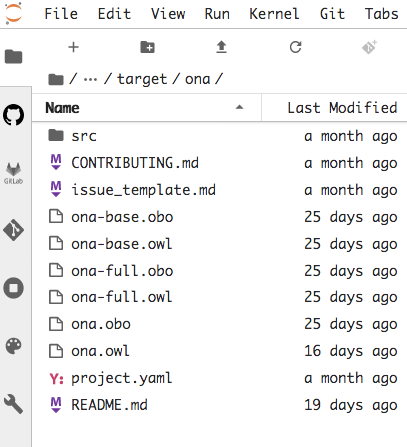
cd target/ona
we have all we need to start developing the ontology under the OBO Foundry guidelines.
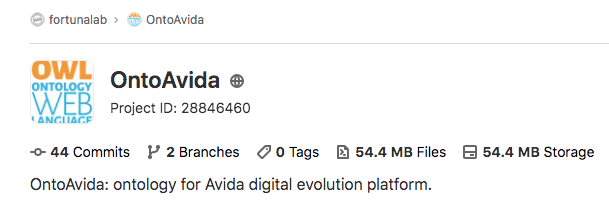
To make the ontology available to the scientific community, we create a repository in GitLab (OntoAvida, as we specified it in the project.yaml file):
We push the project to GitLab:
git remote add origin [email protected]:fortunalab/ontoavida.git
git push -u origin master
Now, everybody can access the ontology tree directory and the ontology OWL file ontoavida.owl.
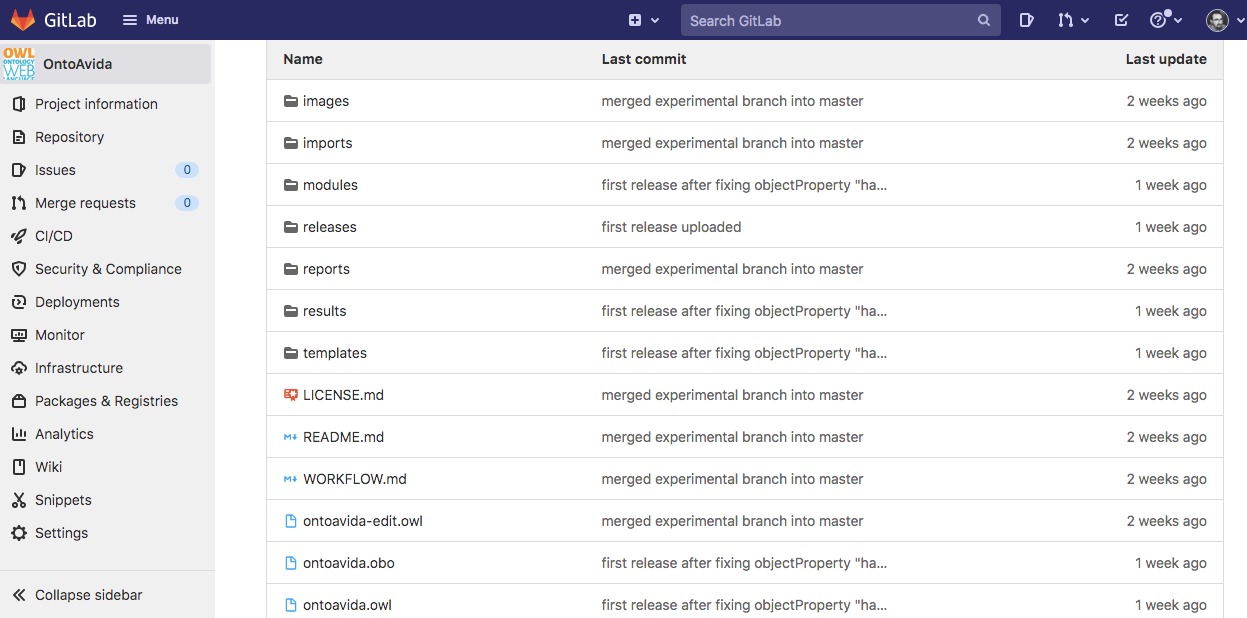
Please use this GitLab repository's Issue tracker to request new terms/classes or report errors or specific concerns related to the ontology.
The Ontology Web Language (OWL) was not designed to be read by humans but by machines (i.e., we are adding semantics to data). That's why the ontoavida.owl file is not reader-friendly. There are software applications specifically developed to visualize and edit owl files. The most widely-used open-source ontology editor is called Protégé, developed by the Stanford Center for Biomedical Informatics Research at the Stanford University School of Medicine.
You can freely download Protégé and then open the ontology file ontoavida.owl to visualize the Classes, Object Properties, and Datatype Properties currently added to the Ontology for Avida.
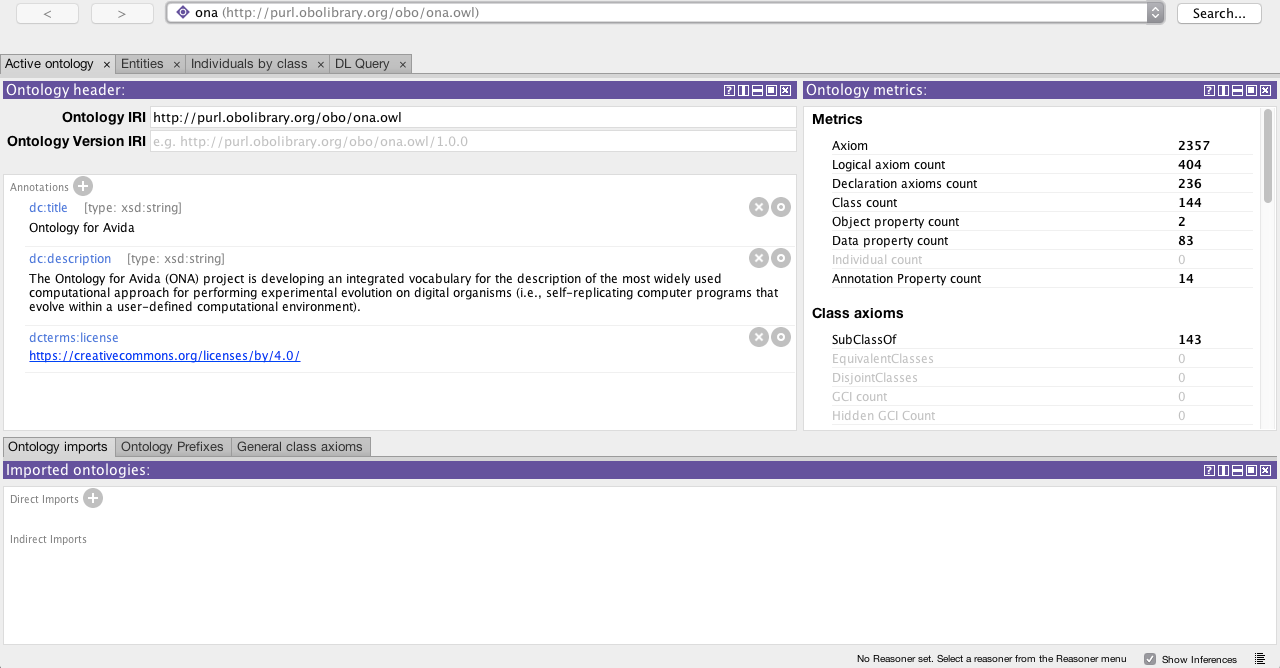
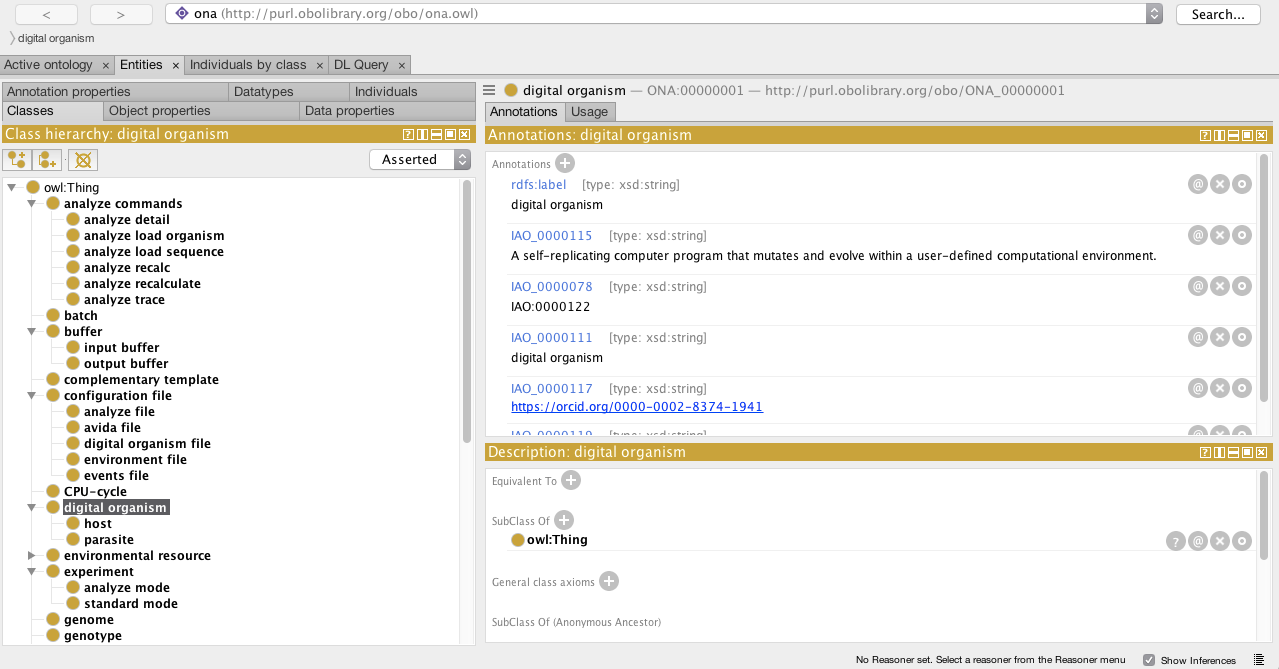
I hope you enjoy it.

